In a new study, Deepmind and colleagues at Isomorphic Labs show early results from a new version of AlphaFold that brings fully automated structure prediction of biological molecules closer to reality.
The Google Deepmind AlphaFold and Isomorphic Labs team today unveiled the latest AlphaFold model. According to the companies, the updated model can now predict the structure of almost any molecule in the Protein Data Bank (PDB), often with atomic accuracy. This development, they say, is an important step towards a better understanding of the complex biological mechanisms within cells.
Since its launch in 2020, AlphaFold has influenced protein structure prediction worldwide. The latest version of the model goes beyond proteins to include a wide range of biologically relevant molecules such as ligands, nucleic acids and post-translational modifications. These structures are critical to understanding biological mechanisms in cells and have been difficult to predict with high accuracy, according to Deepmind.
Deepmind's AlphaFold outperforms specialized prediction models
In predicting protein-ligand interactions, AlphaFold outperforms conventional methods by around 20 percent and can also make predictions for entirely new proteins that have not yet been structurally characterized. Accuracy for protein-protein interfaces has also improved, in some cases significantly, over previous versions, particularly in certain categories such as antibody binding structures.
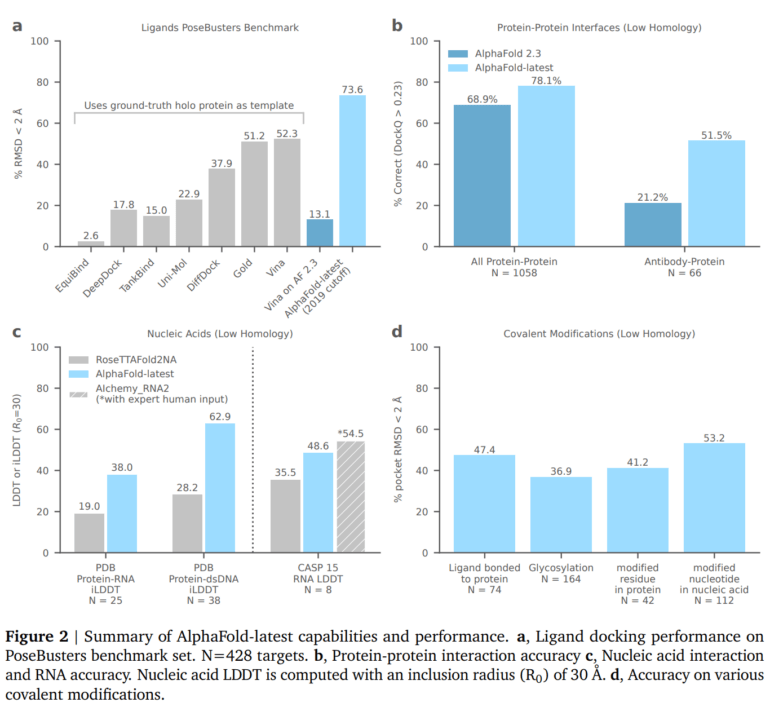
For protein-nucleic acid interfaces, the new version of AlphaFold outperforms competing systems such as RoseTTAFold2NA, while for RNA structure prediction it outperforms other automated methods but is slightly behind the best CASP15 participants using manual intervention by human experts. The improved AlphaFold model could therefore have a significant impact on drug discovery, where the prediction of relevant structures such as antibody binding and protein-ligand binding is important.
New version of AlphaFold brings fully automated structure prediction closer
Although the model presented is still under active development, the team believes that it already demonstrates that computational structure prediction of general biomolecules is possible using machine learning models such as AlphaFold, and opens up many exciting future research avenues for understanding biology.
The goal of fully automated structure prediction directly from sequence is now a step closer.