OpenAI and Ginkgo Bioworks build an autonomous lab where GPT-5 calls the shots

OpenAI, together with biotech company Ginkgo Bioworks, has connected GPT-5 to an automated laboratory to optimize cell-free protein synthesis. The results are measurable, but the limitations are significant.
In fields like math or physics, ideas can be verified through computation alone. Biology works differently. Here, progress runs through the lab, where experiments cost time and money. This is where a new project by OpenAI and biotech company Ginkgo Bioworks comes in.
The two companies connected OpenAI's language model GPT-5 to a fully automated cloud laboratory. The goal was to optimize so-called cell-free protein synthesis (CFPS). In this process, proteins are produced without growing living cells. Instead, the protein production machinery from inside cells is transferred into a controlled mixture and operated there. The method plays a role in drug development, diagnostics, and industrial enzyme production. According to the accompanying paper, it is also used in the commercial manufacturing of an antibody-drug conjugate.
Optimizing CFPS is considered difficult. The mixtures consist of many interacting components, including DNA templates, cell extracts (called lysates, which contain the cellular machinery), energy sources, salts, and cofactors. The space of possible compositions is vast, and the effects of small changes are nearly impossible to predict by intuition alone. Previous attempts using machine learning yielded only incremental improvements.
Six rounds, 36,000 reactions, 40 percent lower costs
Over six iterative rounds, the system tested more than 36,000 different reaction compositions across 580 automated microtiter plates, according to the paper. These are small plastic plates with hundreds of tiny wells, each of which can run a single reaction in parallel. The specific production cost for the test protein sfGFP (a fluorescent standard protein used as a benchmark) dropped from $698 per gram to $422 per gram. That represents a 40 percent reduction compared to the previous state of the art, published by researchers at Northwestern University in August 2025.
At the same time, protein yield increased by 27 percent (from 2.39 to 3.04 grams per liter of reaction solution). Reagent costs alone dropped by 57 percent, from $60 to $26 per gram.
For comparison, the authors cite the list price of a commercially available CFPS kit from NEB, which runs around $800,000 per gram. However, they acknowledge that the figures are not directly comparable.
GPT-5 designs, robots pipette
The process follows a closed loop. GPT-5 generates experimental designs as digital files. A validation system based on the Python library Pydantic automatically checks whether the designed experiments are scientifically sound and physically executable on the available lab automation. Only then are the designs handed off to Ginkgo Bioworks' cloud laboratory in Boston.
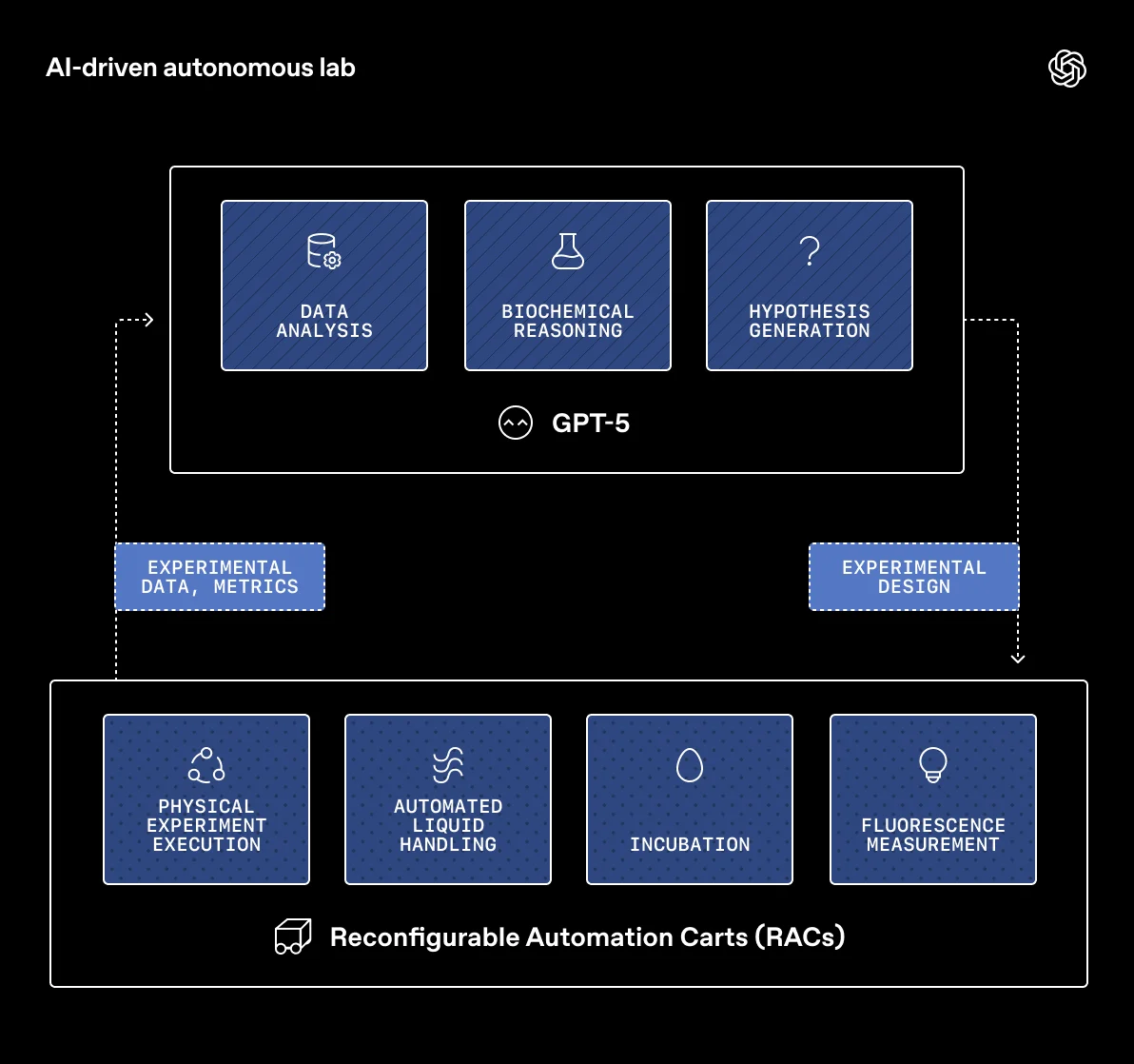
There, modular robotic units called Reconfigurable Automation Carts (RACs) carry out the experiments automatically. Each cart contains a single piece of lab equipment, such as a liquid handler, an incubator, or a measurement instrument. Robotic arms and transport rails move sample plates between stations. Ginkgo's Catalyst software controls the workflow.
Once experiments are complete, the measurement data flows back to GPT-5, which analyzes it, formulates hypotheses, and designs the next round of experiments. According to the authors, human involvement was limited to preparation and loading and unloading of reagents and consumables. In total, the system generated roughly 150,000 data points over six months.
In the first round, GPT-5 designed reaction compositions without any prior example data or experimental results, relying solely on knowledge stored in its weights. Even in this so-called "zero-shot" mode, it produced usable, if not yet optimal, designs.
Tool access drove the biggest gains
Early on, the system struggled with significant variability in measurement results. Deviations between replicates on the same plate sometimes exceeded 40 percent. To improve precision, Ginkgo staff had to manually adjust reagent concentrations and stock solutions. This brought deviations down to a median of 17 percent. The process, in other words, was not purely autonomous.
The biggest performance jump came starting in the third round. At that point, GPT-5 gained access for the first time to a computer, the internet, data analysis packages, and a recent preprint of the previous best results from the Northwestern University researchers. The model also received expanded metadata, including raw data, liquid handling error reports, and actual incubation times.
From round 3 onward, GPT-5 was able to combine its own experimental results with insights from the scientific literature. In just two months (rounds 3 through 5), the system surpassed the previous state of the art. However, the DNA template and cell lysate were also improved at the same time, making it difficult to attribute the gains precisely.
GPT-5 anticipated findings from the field
GPT-5 suggested reagents such as nucleoside monophosphates (NMPs), potassium phosphate, and ribose before it even had access to the preprint mentioned above. The authors of that publication had independently identified the same substances as critical. The model, in other words, reached similar conclusions as domain experts based on its training data and ongoing experimental results alone.
GPT-5 also wrote human-readable lab notebook entries documenting its data analyses and hypotheses. Among other things, the model found that an inexpensive buffer (HEPES) has a disproportionately large effect on protein yield, that phosphate must be buffered within a narrow concentration and pH range, and that adding spermidine (a naturally occurring substance that stabilizes nucleic acids) boosts yields.
The model also made an economic observation. The cost of a CFPS reaction is now more than 90 percent determined by cell lysate and DNA. Increasing protein yield per unit of these expensive ingredients is therefore the most effective lever for cost reduction, rather than savings on cheaper secondary components.
Of the more than 20 additional reagents GPT-5 suggested, several ended up in the best-performing reaction compositions, including NMPs, glucose, potassium phosphate, and catalase.
Few errors, but many open questions
The error rate was low. Of 480 designed microtiter plates, only two had fundamental design flaws, less than one percent. In one case, the model overwrote a prescribed volume specification to make room for additional reagents. In the other, a coding error in unit conversion produced a plate containing only glucose and ribose, which consequently produced no protein.
The project's limitations, however, are substantial. All results come from a single protein (sfGFP) and a single CFPS system. Whether the identified reaction compositions transfer to other proteins remains unclear. In a test with twelve additional proteins, only six were detectable by gel electrophoresis (a common detection method). Further optimization would be needed for other target proteins. Human oversight also remained necessary throughout for protocol improvements and reagent handling.
OpenAI and Ginkgo Bioworks say they plan to extend the approach to additional biological processes. The fact that AI models can independently improve protocols in the wet lab, however, also raises questions. OpenAI points to its own Preparedness Framework for evaluating such capabilities with respect to biosecurity risks. The paper does not name any concrete measures beyond this statement of intent.
AI News Without the Hype – Curated by Humans
As a THE DECODER subscriber, you get ad-free reading, our weekly AI newsletter, the exclusive "AI Radar" Frontier Report 6× per year, access to comments, and our complete archive.
Subscribe nowAI news without the hype
Curated by humans.
- Over 20 percent launch discount.
- Read without distractions – no Google ads.
- Access to comments and community discussions.
- Weekly AI newsletter.
- 6 times a year: “AI Radar” – deep dives on key AI topics.
- Up to 25 % off on KI Pro online events.
- Access to our full ten-year archive.
- Get the latest AI news from The Decoder.