Google DeepMind releases AlphaFold 3 for scientific use
Update –
- Added AlphaFold 3 release
- Added a section about restrictions on accessing AlphaFold 3 compared to AlphaFold 2
Update from November 11, 2024:
Google DeepMind has released AlphaFold 3 on GitHub. The release includes both code and model parameters. According to AlphaFold 3's terms of use for the model parameters, they can be used for non-commercial purposes, particularly research.
The AlphaFold 3 model parameters and output are only available for non-commercial use by, or on behalf of, non-commercial organizations (i.e., universities, non-profit organizations and research institutes, educational, journalism and government bodies). If you are a researcher affiliated with a non-commercial organization, provided you are not a commercial organisation or acting on behalf of a commercial organisation, this means you can use these for your non-commercial affiliated research.
This move addresses earlier criticism that AlphaFold 3's usage was limited to Google DeepMind's server infrastructure (see below).
Update from May 9, 2024:
Access to Alphafold 3 has some restrictions compared to Alphafold 2. Researchers can only use the tool through a server and for non-commercial purposes. They are limited to ten (Update November 2024: 20) predictions per day and cannot calculate the structures of proteins associated with potential drugs.
DeepMind justifies the restrictions by saying that its subsidiary Isomorphic Labs uses Alphafold 3 itself for drug discovery.
"We have to strike a balance between making sure that this is accessible and has the impact in the scientific community as well as not compromising Isomorphic’s ability to pursue commercial drug discovery," Pushmeet Kohli, DeepMind's head of AI science and a co-author of the study, tells Nature.
But it also fuels concerns that DeepMind could monopolize access to the technology and slow scientific progress for commercial interests. Alphafold 2 is open source.
Original article from May 08, 2024:
Google Deepmind unveils AlphaFold 3, the next generation of its protein prediction model
Google DeepMind and its AI medicine spin-off Isomorphic Labs have introduced AlphaFold 3, the latest version of their protein prediction model.
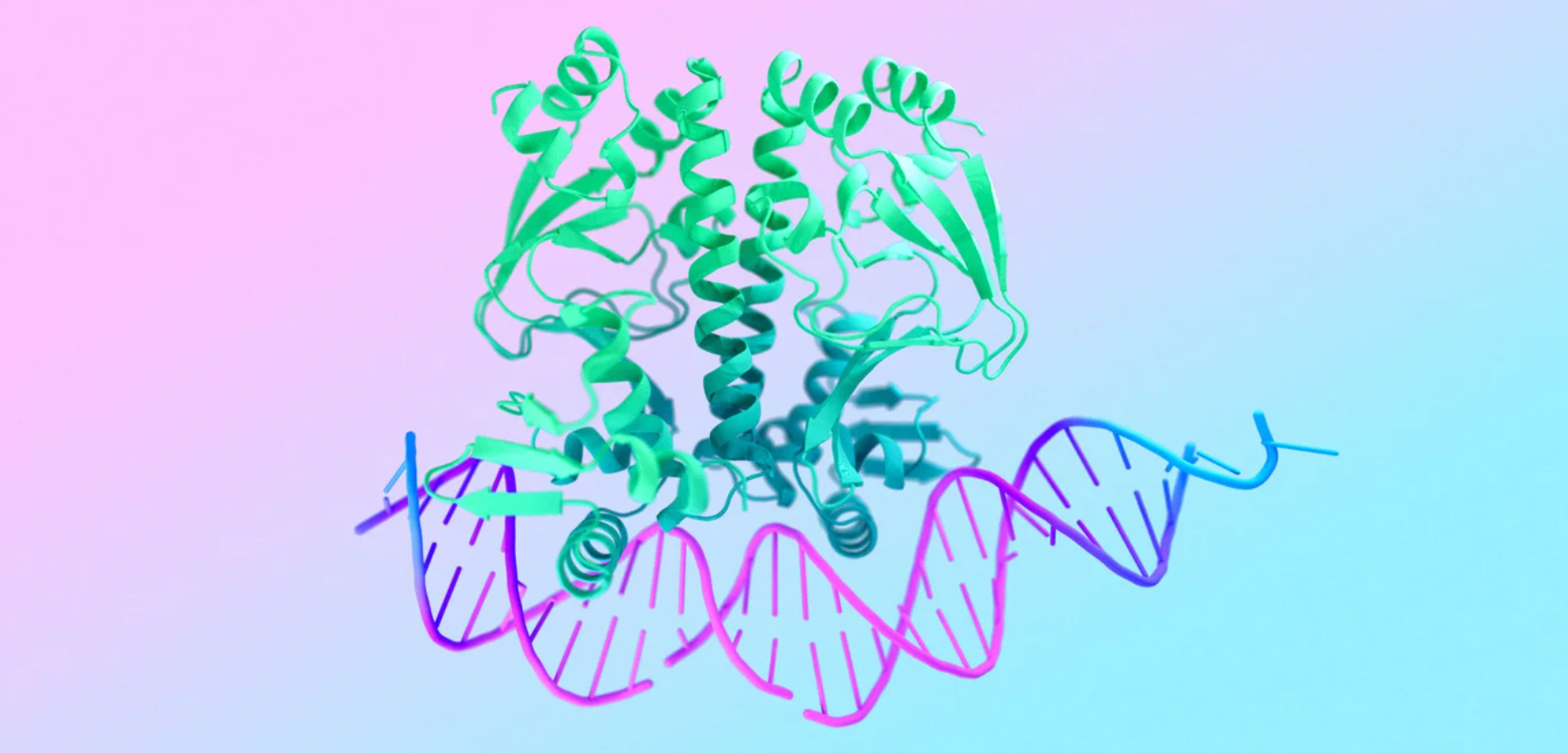
According to Google DeepMind, the new system can predict the structure and interactions of all biological molecules with unprecedented accuracy. It is a major advance over its predecessor, AlphaFold 2, which in 2020 marked a widely recognized breakthrough in protein structure prediction.
While AlphaFold 2 could only predict protein structures, AlphaFold 3 goes much further by modeling the interaction of proteins, DNA, RNA, ligands, ions and chemical modifications. When predicting protein-ligand interactions, AlphaFold 3 is 50 percent more accurate than previous methods, Google DeepMind said.
The core of the model is an improved version of the Evoformer module on which AlphaFold 2 was based, Google DeepMind explained. The final structure prediction uses a new diffusion network, similar to AI image generators.
It starts with a "cloud" of atoms whose positions are randomly noisy, and then gradually tries to reduce the noise and reconstruct the true arrangement of the atoms. This process is iterative, with the model getting a clearer picture of the molecular structure with each step until the final prediction is made at the end.
According to Google DeepMind, what is particularly promising for the development of new drugs is that AlphaFold 3 can predict the interaction of proteins with drugs and antibodies without relying on experimental structural data.
This means that AlphaFold 3 outperforms methods based on physical models that require the exact 3D structure of the protein as a starting point. This in turn must first be determined experimentally, for example by X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy, a time-consuming and costly process. Growing protein crystals for X-ray crystallography can take weeks to months, and NMR experiments require costly equipment and often extensive optimization.
DeepMind is making most of the functions of AlphaFold 3 available for free via the newly launched AlphaFold Server for non-commercial research. With just a few clicks, scientists can use AlphaFold 3, regardless of their experience with machine learning or available computing resources.
The DeepMind team believes that AlphaFold 3 has the potential to "transform our understanding of the biological world and drug discovery." The system "brings the biological world into high definition" and "allows scientists to see cellular systems in all their complexity, across structures, interactions and modifications."
The goal is to accelerate the development of life-changing therapies. However, the benefits still have to be shown in practice. Isomorphic Labs, a DeepMind spin-off, is already using AlphaFold 3 for real-life drug design challenges, both for internal projects and with pharma companies.
AI News Without the Hype – Curated by Humans
As a THE DECODER subscriber, you get ad-free reading, our weekly AI newsletter, the exclusive "AI Radar" Frontier Report 6× per year, access to comments, and our complete archive.
Subscribe nowAI news without the hype
Curated by humans.
- Over 20 percent launch discount.
- Read without distractions – no Google ads.
- Access to comments and community discussions.
- Weekly AI newsletter.
- 6 times a year: “AI Radar” – deep dives on key AI topics.
- Up to 25 % off on KI Pro online events.
- Access to our full ten-year archive.
- Get the latest AI news from The Decoder.