EvolutionaryScale showcases AI that simulates hundreds of millions of years of protein evolution

A new AI model called ESM3 can generate functional proteins that would take hundreds of millions of years to evolve in nature by training on data generated by evolution.
Researchers at EvolutionaryScale have created a large-scale AI model called ESM3 that can generate new functional proteins by training on data produced by evolution. The model can simulate the evolution of proteins and create novel proteins in the process. In a demonstration, ESM3 generated a green fluorescent protein (GFP) called esmGFP, which has only a 58% sequence similarity to the closest known fluorescent protein.
ESM3 was trained on a vast dataset consisting of 2.78 billion natural protein sequences, 236 million protein structures, and 539 million proteins with functional annotations. In total, the model processed 771 billion tokens during training.
Video: EvolutionaryScale
ESM3 processes three-dimensional structures of proteins better than older models
Unlike traditional language models that only learn from textual data, ESM3 learns from discrete tokens representing the sequence, three-dimensional structure, and biological function of proteins. The model views proteins as existing in an organized space where each protein is adjacent to every other protein that differs by a single mutation event.
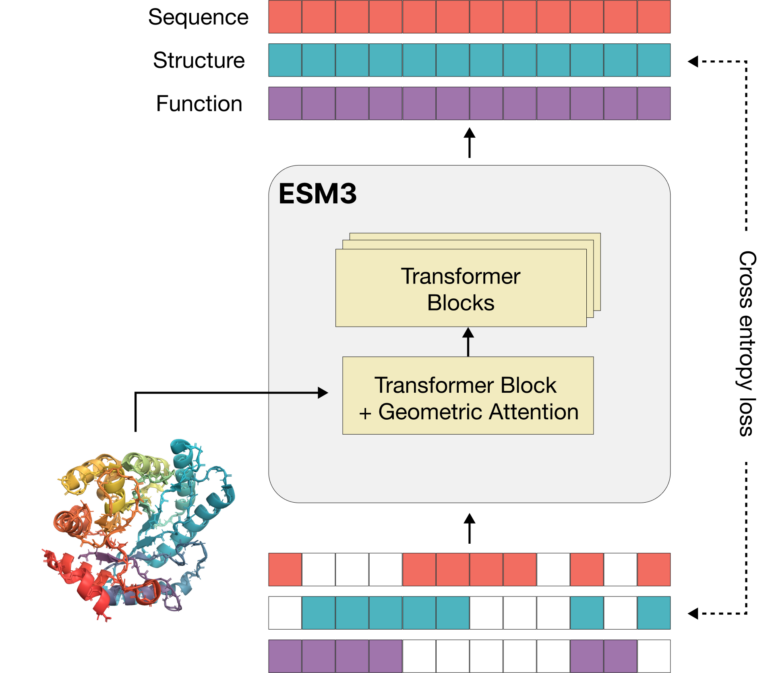
ESM3 uses a novel architecture with "geometric attention" to efficiently process the three-dimensional structure of proteins. This allows the model to understand the organized space of proteins and implicitly construct a model of the many possible evolutionary pathways connecting all proteins without losing the function of the higher-level system.
ESM3 skips 500 million years of evolution
The researchers demonstrated ESM3's ability to generate completely new functional proteins by providing the model with the sequence and structure of key residues that determine fluorescence. Based on this information, ESM3 gradually generated the rest of the protein sequence and structure.
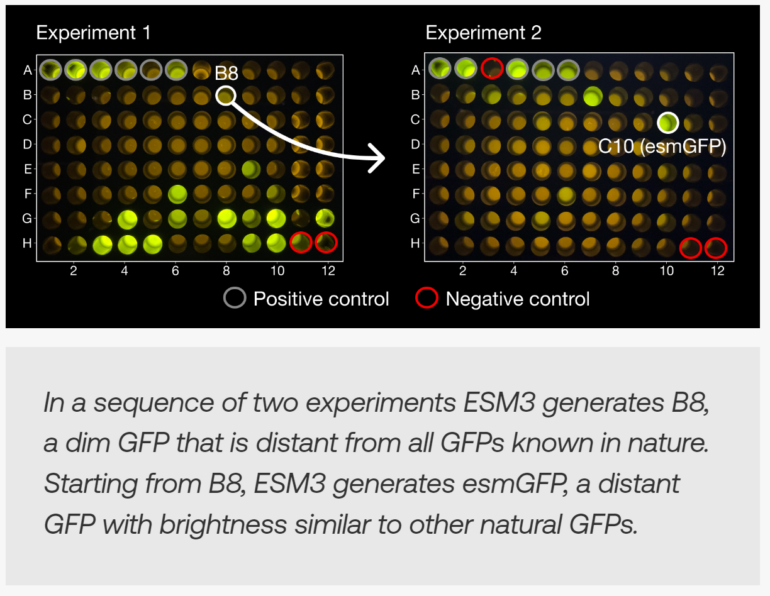
The resulting esmGFP protein has high luminosity despite differing by 58% in its amino acid sequence from the closest known fluorescent protein. Such a significant change would have taken over 500 million years to occur naturally, the team said.
EvolutionaryScale was founded by former meta-researchers
The study once again demonstrates the potential of transformers to capture the biological complexity of proteins and generate new functions. The company's founders have demonstrated this before: They are former members of the Meta-FAIR protein group and were involved in ESMFold, among other projects. Meta disbanded the department in August 2023, while others like Alphabet continue to work in this field with Deepmind's AlphaFold 3.
According to the EvolutionaryScale team, ESM3 now enables a program-driven approach to protein design that bridges the gap between human specifications and the complexity of biology. In the future, this technique could enable numerous applications in biotechnology and medicine.
However, the researchers also emphasize the need for responsible use of such powerful AI models. They are therefore releasing an open version of ESM3 for researchers to use. According to the team, the model has been tested for safety by experts. According to the experts, the positive effects of the release outweigh the risks.
The complete ESM3 models are to be made available via an API with free access for academic research.
AI News Without the Hype – Curated by Humans
As a THE DECODER subscriber, you get ad-free reading, our weekly AI newsletter, the exclusive "AI Radar" Frontier Report 6× per year, access to comments, and our complete archive.
Subscribe nowAI news without the hype
Curated by humans.
- Over 20 percent launch discount.
- Read without distractions – no Google ads.
- Access to comments and community discussions.
- Weekly AI newsletter.
- 6 times a year: “AI Radar” – deep dives on key AI topics.
- Up to 25 % off on KI Pro online events.
- Access to our full ten-year archive.
- Get the latest AI news from The Decoder.